- Home
- Lab-services
- Sequence-check-tools
Sequence check tools
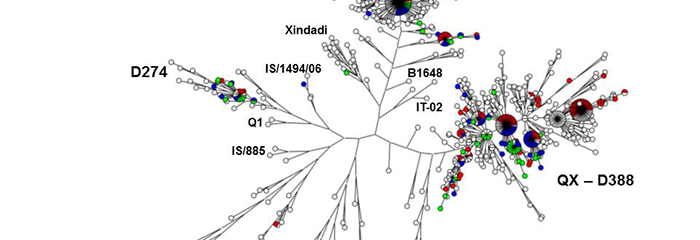
Worldwide, more and more viruses and bacteria are detected by means of PCR and subsequently genotyped using DNA sequencing. DNA sequencing is more affordable nowadays and is offered by an increasing number of laboratories. As a result of this, the number of sequences in GenBank® will keep on growing.
A major challenge is the interpretation of the detected sequences. Many laboratories report unfamiliar genetic codes or sequences, but you need to know what this means. Is the detected sequence from a vaccine strain or is it a field strain, and if so, which strain? You need to know the biological relevance of these strains. We offer several sequence check tools to help you with the interpretation of your sequencing results or raw sequence data:
- Sequence Check - Virus ID
- Sequence Check - FASTA file
- Sequence Check - Sanger sequence reads
- Sequence Check - Molecular epidemiological analysis

Virus ID
You provide the reported virus name or code and we help you determine the genotype and supply information about the similarity to known reference genotypes. You will receive your sequence of interest in a phylogenetic tree containing relevant genotypes available in our database.
FASTA file
You provide a FASTA file containing the DNA sequence of your strain of interest because you want to determine the genotype and receive information about the similarity to known reference genotypes. You will receive your sequence of interest in a phylogenetic tree containing relevant genotypes available in our database.
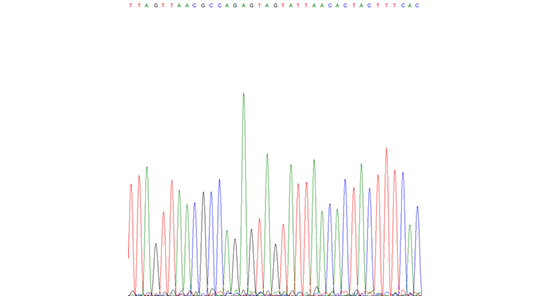
Sanger sequence reads
You provide the raw sequence reads of your samples because you want to determine the genotype and receive information about the similarity to known reference genotypes. In addition, if your sample contains a mixture of two strains or an unknown genotype, we can help you determine which strains are involved. You will receive your sequence of interest in a phylogenetic tree containing relevant genotypes available in our database.
Molecular epidemiological analysis
You provide the sample IDs of samples previously sequenced for you by GD. This can be, for instance, sequences of samples collected from different flocks at the same farm or sequences of strains collected at different farms over time. You will receive a phylogenetic tree to show the genetic relatedness of your samples, compared with each other and compared with relevant genotypes available in our database. Alternatively, you can provide FASTA files or Sanger sequence reads of samples sequenced by other labs.
Order now
You can order the GD Sequence Check tools online by filling out the submission form. If you have any further questions, please do not hesitate to contact us via +31 (0)88 20 25 575 or by email.